
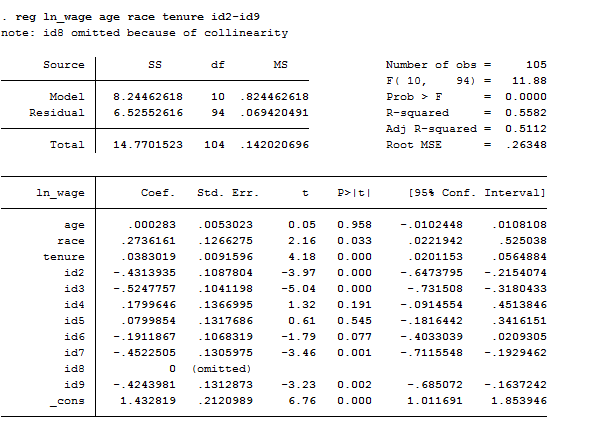
Mi estimate, saving(myestimates, replace). If you're planning to do postestimation, tell mi estimate to store the needed information in a small file with the saving() option: Some tasks require the e() vector from the regression run on each completed data set. Unlike standard estimation commands, mi estimate cannot save all the information needed for postestimation tasks in the e() vector. See White, Royston, and Wood for a list of quantities that can and cannot be combined using Rubin's Rules. Others simply cannot, such as likelihood ratio test statistics. Some quantities can be estimated if they are transformed to make them approximately normal, such as R-squared values. Fortunately, regression coefficients do meet those assumptions. Rubin's rules require certain assumptions to be valid, notably asymptotic normality, and if a quantity does not meet those assumptions then Rubin's rules cannot provide a valid estimate of it. Postestimation with imputed data must be done with caution. In that case specifying the base category should fix the problem. But it can also arise from the estimation command choosing different base categories. In our experience that's been the result of perfect prediction in some imputations and not others, which suggests problems with the model being run (such as too many categorical covariates for the number of observations available). More rarely, you could run into problems with different imputations using different sets of variables. Of course this raises the same issues as complete cases analysis, though the effects will likely be smaller. Mi estimate: reg wage edu exp if race=1 & !miss_race If so, you can use those variables as part of your subsample selection: Hopefully you created indicator variables telling you which observations are missing which variables in the process of determining whether your data are MCAR, MCAR or MNAR with: The other is to not use observations that have imputed values of the variables used to select the subsample. Mi estimate, esampvaryok: reg wage edu exp if race=1 The Stata documentation says this may result in "may result in biased or inefficient estimates" but we don't have any guidance at this time as to the seriousness of the problem. One is to simply tell mi estimate to ignore the problem with the esampvaryok option.


Thus the subsample to be used will vary between imputations, and mi estimate will give you an error message. If race is an imputed variable, then some observations will likely have a one for race in some imputations and not others. However, it is your responsibility to ensure that the results will be valid. If a command is not on that list, you can tell mi estimate to apply them anyway with the cmdok ("command ok") option. Mi estimate has a list of estimation commands for which it knows Rubin's rules are appropriate. Because the output is created by mi estimate, options that affect output, such as or to display odds ratios, must be applied to mi estimate rather than the estimation command.
It then combines the results using Rubin's rules and displays the output. The mi estimate command first runs the estimation command on each imputation separately. It is a prefix command, like svy or by, meaning that it goes in front of whatever estimation command you're running. The main command for running estimations on imputed data is mi estimate. Once the imputations are created and checked, Stata makes estimation using the imputed data relatively easy. In most cases, the hard work of using multiple imputation comes in the imputation process. For a list of topics covered by this series, see the Introduction. This article is part of the Multiple Imputation in Stata series.


 0 kommentar(er)
0 kommentar(er)
